Ice thickness inversion#
This example shows how to run the OGGM ice thickness inversion model with various ice parameters: the deformation parameter A and a sliding parameter (fs).
There is currently no “best” set of parameters for the ice thickness inversion model. As shown in Maussion et al. (2019), the default parameter set results in global volume estimates which are a bit larger than previous values. For the consensus estimate of Farinotti et al. (2019), OGGM participated with a deformation parameter A that is 1.5 times larger than the generally accepted default value.
There is no reason to think that the ice parameters are the same between neighboring glaciers. There is currently no “good” way to calibrate them, or at least no generaly accepted one. We won’t discuss the details here, but we provide a script to illustrate the sensitivity of the model to this choice.
New in version 1.4: we demonstrate how to apply a new global task in OGGM, workflow.calibrate_inversion_from_consensus to calibrate the A parameter to match the consensus estimate from Farinotti et al. (2019).
At the end of this tutorial, we show how to distribute the “flowline thicknesses” on a glacier map.
Run#
# Libs
import geopandas as gpd
# Locals
import oggm.cfg as cfg
from oggm import utils, workflow, tasks, graphics
# Initialize OGGM and set up the default run parameters
cfg.initialize(logging_level='WARNING')
rgi_region = '11' # Region Central Europe
# Local working directory (where OGGM will write its output)
WORKING_DIR = utils.gettempdir('OGGM_Inversion')
cfg.PATHS['working_dir'] = WORKING_DIR
# This is useful here
cfg.PARAMS['use_multiprocessing'] = True
# RGI file
path = utils.get_rgi_region_file(rgi_region)
rgidf = gpd.read_file(path)
# Select the glaciers in the Pyrenees
rgidf = rgidf.loc[rgidf['O2Region'] == '2']
# Sort for more efficient parallel computing
rgidf = rgidf.sort_values('Area', ascending=False)
# Go - get the pre-processed glacier directories
# We start at level 3, because we need all data for the inversion
# we need centerlines to use all functions of the distributed ice thickness later
# (we specifically need `geometries.pkl` in the gdirs)
cfg.PARAMS['border'] = 80
base_url = ('https://cluster.klima.uni-bremen.de/~oggm/gdirs/oggm_v1.6/'
'L3-L5_files/2023.3/centerlines/W5E5/')
gdirs = workflow.init_glacier_directories(rgidf, from_prepro_level=3,
prepro_base_url=base_url)
# to assess the model geometry (for e.g. distributed ice thickness plots),
# we need to set that to true
#cfg.PARAMS['store_model_geometry'] = True
#workflow.execute_entity_task(tasks.prepare_for_inversion, gdirs)
# Default parameters
# Deformation: from Cuffey and Patterson 2010
glen_a = 2.4e-24
# Sliding: from Oerlemans 1997
fs = 5.7e-20
2024-04-25 13:29:42: oggm.cfg: Reading default parameters from the OGGM `params.cfg` configuration file.
2024-04-25 13:29:42: oggm.cfg: Multiprocessing switched OFF according to the parameter file.
2024-04-25 13:29:42: oggm.cfg: Multiprocessing: using all available processors (N=4)
2024-04-25 13:29:43: oggm.cfg: Multiprocessing switched ON after user settings.
2024-04-25 13:30:16: oggm.workflow: init_glacier_directories from prepro level 3 on 35 glaciers.
2024-04-25 13:30:16: oggm.workflow: Execute entity tasks [gdir_from_prepro] on 35 glaciers
with utils.DisableLogger(): # this scraps some output - to use with caution!!!
# Correction factors
factors = [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1]
factors += [1.1, 1.2, 1.3, 1.5, 1.7, 2, 2.5, 3, 4, 5]
factors += [6, 7, 8, 9, 10]
# Run the inversions tasks with the given factors
for f in factors:
# Without sliding
suf = '_{:03d}_without_fs'.format(int(f * 10))
workflow.execute_entity_task(tasks.mass_conservation_inversion, gdirs,
glen_a=glen_a*f, fs=0)
# Store the results of the inversion only
utils.compile_glacier_statistics(gdirs, filesuffix=suf,
inversion_only=True)
# With sliding
suf = '_{:03d}_with_fs'.format(int(f * 10))
workflow.execute_entity_task(tasks.mass_conservation_inversion, gdirs,
glen_a=glen_a*f, fs=fs)
# Store the results of the inversion only
utils.compile_glacier_statistics(gdirs, filesuffix=suf,
inversion_only=True)
Read the data#
The data are stored as csv files in the working directory. The easiest way to read them is to use pandas!
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from scipy import stats
import os
Let’s read the output of the inversion with the default OGGM parameters first:
df = pd.read_csv(os.path.join(WORKING_DIR, 'glacier_statistics_011_without_fs.csv'), index_col=0)
df
| rgi_region | rgi_subregion | name | cenlon | cenlat | rgi_area_km2 | rgi_year | glacier_type | terminus_type | is_tidewater | ... | vas_volume_km3 | inv_volume_bsl_km3 | inv_volume_bwl_km3 | dem_source | n_orig_centerlines | flowline_type | perc_invalid_flowline | apparent_mb_from_any_mb_residual | inversion_glen_a | inversion_fs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rgi_id | |||||||||||||||||||||
| RGI60-11.03208 | 11 | 11-02 | Aneto | 0.646032 | 42.641357 | 0.622 | 2011 | Glacier | Land-terminating | False | ... | 0.017699 | 0.0 | 0.0 | NASADEM | 2 | centerlines | 0.205128 | 737.400000 | 2.640000e-24 | 0 |
| RGI60-11.03232 | 11 | 11-02 | Ossoue | -0.141222 | 42.771191 | 0.449 | 2011 | Glacier | Land-terminating | False | ... | 0.011306 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1168.200000 | 2.640000e-24 | 0 |
| RGI60-11.03222 | 11 | 11-02 | Monte Perdido Lower | 0.039655 | 42.679337 | 0.308 | 2011 | Glacier | Land-terminating | False | ... | 0.006733 | 0.0 | 0.0 | NASADEM | 2 | centerlines | 0.000000 | 887.700000 | 2.640000e-24 | 0 |
| RGI60-11.03209 | 11 | 11-02 | Maladeta E | 0.639343 | 42.648979 | 0.260 | 2011 | Glacier | Land-terminating | False | ... | 0.005334 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 593.500000 | 2.640000e-24 | 0 |
| RGI60-11.03231 | 11 | 11-02 | Oulettes Du Gaube | -0.144215 | 42.779716 | 0.130 | 2011 | Glacier | Land-terminating | False | ... | 0.002057 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 509.400000 | 2.640000e-24 | 0 |
| RGI60-11.03213 | 11 | 11-02 | Seil De La Baque E | 0.494304 | 42.694481 | 0.105 | 2011 | Glacier | Land-terminating | False | ... | 0.001533 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 572.600000 | 2.640000e-24 | 0 |
| RGI60-11.03239 | 11 | 11-02 | Jezerces Gl No1 | 19.807627 | 42.465839 | 0.097 | 2007 | Glacier | Land-terminating | False | ... | 0.001375 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.562500 | 731.500000 | 2.640000e-24 | 0 |
| RGI60-11.03229 | 11 | 11-02 | Gabietous | -0.057456 | 42.695129 | 0.087 | 2011 | Glacier | Land-terminating | False | ... | 0.001184 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1600.600000 | 2.640000e-24 | 0 |
| RGI60-11.03218 | 11 | 11-02 | Llardana | 0.428193 | 42.653946 | 0.084 | 2011 | Glacier | Land-terminating | False | ... | 0.001128 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1095.300000 | 2.640000e-24 | 0 |
| RGI60-11.03228 | 11 | 11-02 | Taillon | -0.039683 | 42.695419 | 0.083 | 2011 | Glacier | Land-terminating | False | ... | 0.001110 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1738.800000 | 2.640000e-24 | 0 |
| RGI60-11.03205 | 11 | 11-02 | Tempestades | 0.666092 | 42.626839 | 0.077 | 2011 | Glacier | Land-terminating | False | ... | 0.001001 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 108.800000 | 2.640000e-24 | 0 |
| RGI60-11.03206 | 11 | 11-02 | Barrancs | 0.658785 | 42.632011 | 0.067 | 2011 | Glacier | Land-terminating | False | ... | 0.000827 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1392.800000 | 2.640000e-24 | 0 |
| RGI60-11.03217 | 11 | 11-02 | La Paul | 0.437356 | 42.661072 | 0.057 | 2011 | Glacier | Land-terminating | False | ... | 0.000662 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 882.900000 | 2.640000e-24 | 0 |
| RGI60-11.03221 | 11 | 11-02 | Monte Perdido Upper | 0.033972 | 42.678589 | 0.054 | 2011 | Glacier | Land-terminating | False | ... | 0.000614 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 871.000000 | 2.640000e-24 | 0 |
| RGI60-11.03210 | 11 | 11-02 | Maladeta W | 0.632566 | 42.652107 | 0.054 | 2011 | Glacier | Land-terminating | False | ... | 0.000614 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 402.100000 | 2.640000e-24 | 0 |
| RGI60-11.03233 | 11 | 11-02 | Infierno Central | -0.258849 | 42.783802 | 0.053 | 2011 | Glacier | Land-terminating | False | ... | 0.000599 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 532.200000 | 2.640000e-24 | 0 |
| RGI60-11.03225 | 11 | 11-02 | Astazou | 0.034118 | 42.703136 | 0.052 | 2011 | Glacier | Land-terminating | False | ... | 0.000583 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1260.300000 | 2.640000e-24 | 0 |
| RGI60-11.03211 | 11 | 11-02 | Boum | 0.555278 | 42.700157 | 0.050 | 2011 | Glacier | Land-terminating | False | ... | 0.000553 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 687.600000 | 2.640000e-24 | 0 |
| RGI60-11.03240 | 11 | 11-02 | Jezerces Gl No2 | 19.815450 | 42.447353 | 0.049 | 2007 | Glacier | Land-terminating | False | ... | 0.000538 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1706.700000 | 2.640000e-24 | 0 |
| RGI60-11.03227 | 11 | 11-02 | Pailla W | 0.015287 | 42.705482 | 0.046 | 2011 | Glacier | Land-terminating | False | ... | 0.000493 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 1058.400000 | 2.640000e-24 | 0 |
| RGI60-11.03219 | 11 | 11-02 | Barroude | 0.142224 | 42.726147 | 0.042 | 2011 | Glacier | Land-terminating | False | ... | 0.000435 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 620.000000 | 2.640000e-24 | 0 |
| RGI60-11.03220 | 11 | 11-02 | Munia | 0.129816 | 42.717735 | 0.041 | 2011 | Glacier | Land-terminating | False | ... | 0.000421 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 383.300000 | 2.640000e-24 | 0 |
| RGI60-11.03212 | 11 | 11-02 | Portillon D Oo | 0.511460 | 42.693188 | 0.040 | 2011 | Glacier | Land-terminating | False | ... | 0.000407 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 532.500000 | 2.640000e-24 | 0 |
| RGI60-11.03226 | 11 | 11-02 | Pailla E | 0.021260 | 42.705322 | 0.035 | 2011 | Glacier | Land-terminating | False | ... | 0.000339 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | -763.200000 | 2.640000e-24 | 0 |
| RGI60-11.03241 | 11 | 11-02 | It4L30000001 Gh Del Calderone | 13.566710 | 42.470540 | 0.033 | 1990 | Glacier | Land-terminating | False | ... | 0.000312 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.250000 | 1036.800000 | 2.640000e-24 | 0 |
| RGI60-11.03230 | 11 | 11-02 | Petit Vignemale | -0.137718 | 42.775856 | 0.027 | 2011 | Glacier | Land-terminating | False | ... | 0.000237 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 131.700000 | 2.640000e-24 | 0 |
| RGI60-11.03215 | 11 | 11-02 | Gourgs Blancs | 0.480293 | 42.700859 | 0.026 | 2011 | Glacier | Land-terminating | False | ... | 0.000225 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 677.300000 | 2.640000e-24 | 0 |
| RGI60-11.03234 | 11 | 11-02 | Las Neous | -0.286945 | 42.838642 | 0.025 | 2011 | Glacier | Land-terminating | False | ... | 0.000213 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 346.200000 | 2.640000e-24 | 0 |
| RGI60-11.03223 | 11 | 11-02 | Marbore | 0.018435 | 42.693417 | 0.023 | 2011 | Glacier | Land-terminating | False | ... | 0.000190 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 596.371893 | 2.640000e-24 | 0 |
| RGI60-11.03224 | 11 | 11-02 | Cilindro | 0.026603 | 42.689346 | 0.022 | 2011 | Glacier | Land-terminating | False | ... | 0.000179 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 760.500000 | 2.640000e-24 | 0 |
| RGI60-11.03204 | 11 | 11-02 | Valier | 1.088501 | 42.798626 | 0.021 | 2011 | Glacier | Land-terminating | False | ... | 0.000168 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 596.371893 | 2.640000e-24 | 0 |
| RGI60-11.03214 | 11 | 11-02 | Seil De La Baque W | 0.490498 | 42.696220 | 0.020 | 2011 | Glacier | Land-terminating | False | ... | 0.000157 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 422.800000 | 2.640000e-24 | 0 |
| RGI60-11.03238 | 11 | 11-02 | Debeli Namet Gl | 19.067500 | 43.115000 | 0.019 | 2003 | Glacier | Land-terminating | False | ... | 0.000146 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 596.371893 | 2.640000e-24 | 0 |
| RGI60-11.03207 | 11 | 11-02 | Coronas | 0.654112 | 42.632977 | 0.015 | 2011 | Glacier | Land-terminating | False | ... | 0.000106 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 596.371893 | 2.640000e-24 | 0 |
| RGI60-11.03216 | 11 | 11-02 | Posets | 0.438703 | 42.656570 | 0.010 | 2011 | Glacier | Land-terminating | False | ... | 0.000060 | 0.0 | 0.0 | NASADEM | 1 | centerlines | 0.000000 | 596.371893 | 2.640000e-24 | 0 |
35 rows × 28 columns
There are only 35 glaciers in the Pyrenees! That’s why the run was relatively fast.
Results#
One way to visualize the output is to plot the volume as a function of area in a log-log plot, illustrating the well known volume-area relationship of mountain glaciers:
ax = df.plot(kind='scatter', x='rgi_area_km2', y='inv_volume_km3')
ax.semilogx(); ax.semilogy()
xlim, ylim = [1e-2, 0.7], [1e-5, 0.05]
ax.set_xlim(xlim); ax.set_ylim(ylim);
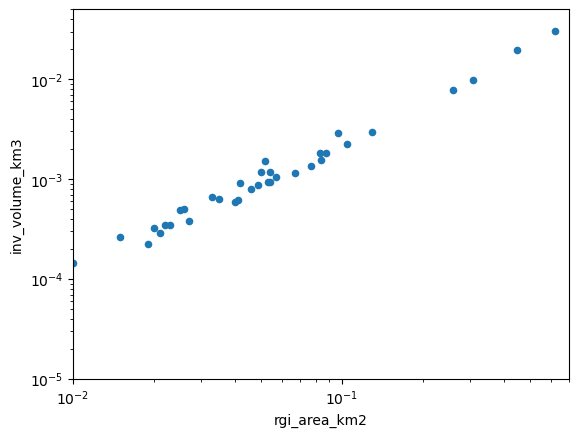
As we can see, there is a clear relationship, but it is not perfect. Let’s fit a line to these data (the “volume-area scaling law”):
# Fit in log space
dfl = np.log(df[['inv_volume_km3', 'rgi_area_km2']])
slope, intercept, r_value, p_value, std_err = stats.linregress(dfl.rgi_area_km2.values, dfl.inv_volume_km3.values)
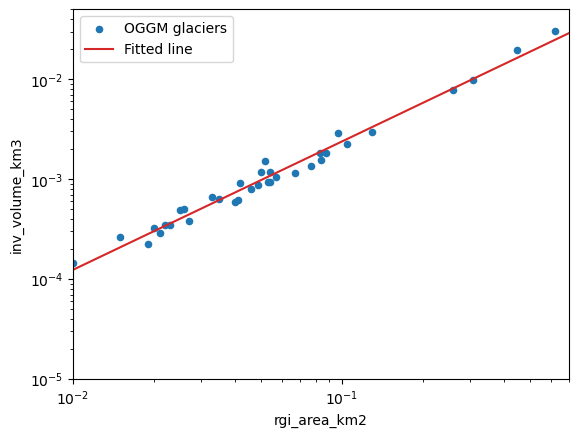
In their seminal paper, Bahr et al. (1997) describe this relationship as:
With V the volume in km\(^3\), S the area in km\(^2\) and \(\alpha\) and \(\gamma\) the scaling parameters (0.034 and 1.375, respectively). How does OGGM compare to these in the Pyrenees?
print('power: {:.3f}'.format(slope))
print('slope: {:.3f}'.format(np.exp(intercept)))
power: 1.284
slope: 0.046
ax = df.plot(kind='scatter', x='rgi_area_km2', y='inv_volume_km3', label='OGGM glaciers')
ax.plot(xlim, np.exp(intercept) * (xlim ** slope), color='C3', label='Fitted line')
ax.semilogx(); ax.semilogy()
ax.set_xlim(xlim); ax.set_ylim(ylim);
ax.legend();
Sensitivity analysis#
Now, let’s read the output files of each run separately, and compute the regional volume out of them:
dftot = pd.DataFrame(index=factors)
for f in factors:
# Without sliding
suf = '_{:03d}_without_fs'.format(int(f * 10))
fpath = os.path.join(WORKING_DIR, 'glacier_statistics{}.csv'.format(suf))
_df = pd.read_csv(fpath, index_col=0, low_memory=False)
dftot.loc[f, 'without_sliding'] = _df.inv_volume_km3.sum()
# With sliding
suf = '_{:03d}_with_fs'.format(int(f * 10))
fpath = os.path.join(WORKING_DIR, 'glacier_statistics{}.csv'.format(suf))
_df = pd.read_csv(fpath, index_col=0, low_memory=False)
dftot.loc[f, 'with_sliding'] = _df.inv_volume_km3.sum()
And plot them:
dftot.plot();
plt.xlabel('Factor of Glen A (default 1)'); plt.ylabel('Regional volume (km$^3$)');
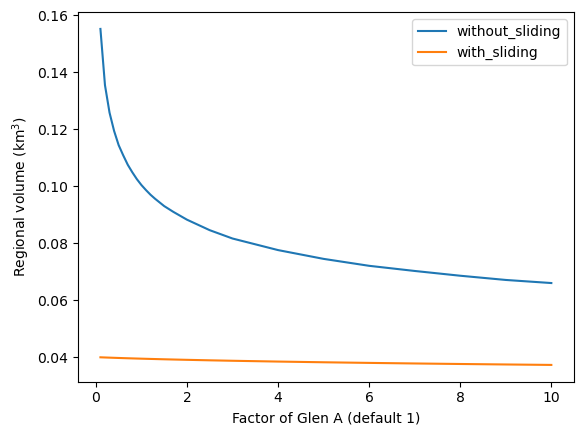
As you can see, there is quite a difference between the solutions. In particular, close to the default value for Glen A, the regional estimates are very sensitive to small changes in A. The calibration of A is a problem that has yet to be resolved by global glacier models…
Since OGGM version 1.4: calibrate to match the consensus estimate#
Here, one “best Glen A” is found in order that the total inverted volume of the glaciers of gdirs fits to the 2019 consensus estimate.
# when we use all glaciers, no Glen A could be found within the range [0.1,10] that would match the consensus estimate
# usually, this is applied on larger regions where this error should not occur !
cdf = workflow.calibrate_inversion_from_consensus(gdirs[1:], filter_inversion_output=False)
2024-04-25 13:35:30: oggm.workflow: Applying global task calibrate_inversion_from_consensus on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Consensus estimate optimisation with A factor: 0.1 and fs: 0
2024-04-25 13:35:30: oggm.workflow: Applying global task inversion_tasks on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [prepare_for_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [mass_conservation_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [get_inversion_volume] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Consensus estimate optimisation with A factor: 10.0 and fs: 0
2024-04-25 13:35:30: oggm.workflow: Applying global task inversion_tasks on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [prepare_for_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [mass_conservation_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [get_inversion_volume] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Consensus estimate optimisation with A factor: 9.628954293399921 and fs: 0
2024-04-25 13:35:30: oggm.workflow: Applying global task inversion_tasks on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [prepare_for_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [mass_conservation_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [get_inversion_volume] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Consensus estimate optimisation with A factor: 7.584273809665188 and fs: 0
2024-04-25 13:35:30: oggm.workflow: Applying global task inversion_tasks on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [prepare_for_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [mass_conservation_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [get_inversion_volume] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Consensus estimate optimisation with A factor: 7.4100273087099495 and fs: 0
2024-04-25 13:35:30: oggm.workflow: Applying global task inversion_tasks on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [prepare_for_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [mass_conservation_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [get_inversion_volume] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Consensus estimate optimisation with A factor: 7.3729771721654 and fs: 0
2024-04-25 13:35:30: oggm.workflow: Applying global task inversion_tasks on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [prepare_for_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [mass_conservation_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [get_inversion_volume] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: calibrate_inversion_from_consensus converged after 5 iterations and fs=0. The resulting Glen A factor is 7.4100273087099495.
2024-04-25 13:35:30: oggm.workflow: Applying global task inversion_tasks on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [prepare_for_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [mass_conservation_inversion] on 34 glaciers
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [get_inversion_volume] on 34 glaciers
cdf.sum()
vol_itmix_m3 4.859110e+07
vol_bsl_itmix_m3 0.000000e+00
vol_oggm_m3 4.858508e+07
dtype: float64
Note that here we calibrate the Glen A parameter to a value that is equal for all glaciers of gdirs, i.e. we calibrate to match the total volume of all glaciers and not to match them individually.
cdf.iloc[:3]
| vol_itmix_m3 | vol_bsl_itmix_m3 | vol_oggm_m3 | |
|---|---|---|---|
| RGIId | |||
| RGI60-11.03232 | 1.422478e+07 | 0.0 | 1.380812e+07 |
| RGI60-11.03222 | 5.158277e+06 | 0.0 | 7.253356e+06 |
| RGI60-11.03209 | 5.335579e+06 | 0.0 | 5.483800e+06 |
just as a side note, “vol_bsl_itmix_m3” means volume below sea level and is therefore zero for these Alpine glaciers!
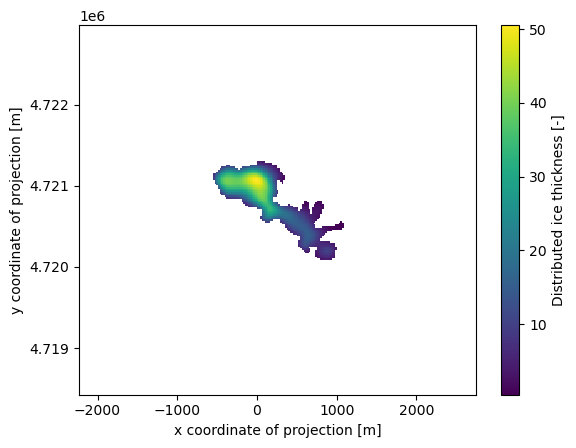
Distributed ice thickness#
The OGGM inversion and dynamical models use the “1D” flowline assumption: for some applications, you might want to use OGGM to create distributed ice thickness maps. Currently, OGGM implements two ways to “distribute” the flowline thicknesses, but only the simplest one works robustly:
# Distribute
workflow.execute_entity_task(tasks.distribute_thickness_per_altitude, gdirs);
2024-04-25 13:35:30: oggm.workflow: Execute entity tasks [distribute_thickness_per_altitude] on 35 glaciers
We just created a new output of the model, which we can access in the gridded_data file:
# xarray is an awesome library! Did you know about it?
import xarray as xr
import rioxarray as rioxr
ds = xr.open_dataset(gdirs[0].get_filepath('gridded_data'))
ds.distributed_thickness.plot();
Since some people find geotiff data easier to read than netCDF, OGGM also provides a tool to convert the variables in gridded_data.nc file to a geotiff file:
# save the distributed ice thickness into a geotiff file
workflow.execute_entity_task(tasks.gridded_data_var_to_geotiff, gdirs, varname='distributed_thickness')
# The default path of the geotiff file is in the glacier directory with the name "distributed_thickness.tif"
# Let's check if the file exists
for gdir in gdirs:
path = os.path.join(gdir.dir, 'distributed_thickness.tif')
assert os.path.exists(path)
2024-04-25 13:35:32: oggm.workflow: Execute entity tasks [gridded_data_var_to_geotiff] on 35 glaciers
# Open the last file with xarray's open_rasterio
rioxr.open_rasterio(path).plot();
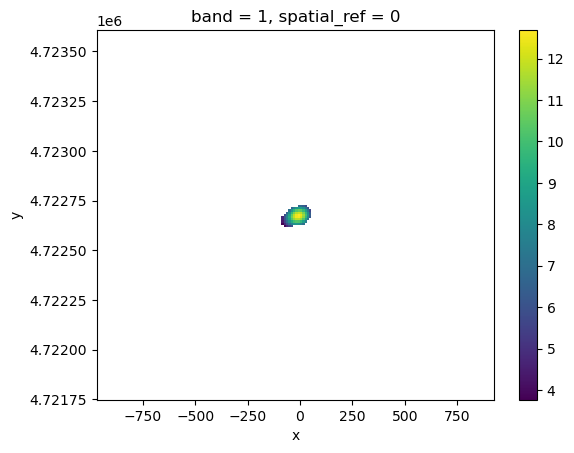
In fact, tasks.gridded_data_var_to_geotiff() can save any variable in the gridded_data.nc file. The geotiff is named as the variable name with a .tif suffix. Have a try by yourself ;-)
Plot many glaciers on a map#
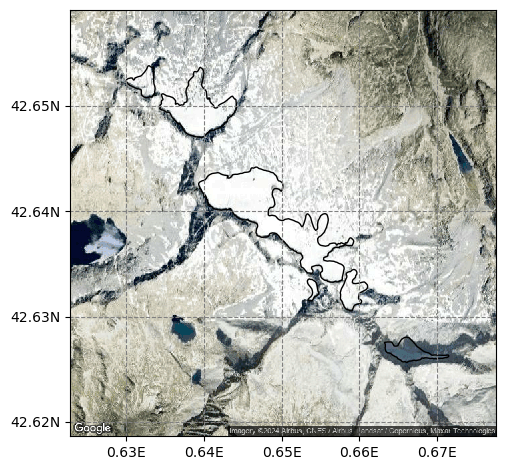
Let’s select a group of glaciers close to each other:
rgi_ids = ['RGI60-11.0{}'.format(i) for i in range(3205, 3211)]
sel_gdirs = [gdir for gdir in gdirs if gdir.rgi_id in rgi_ids]
graphics.plot_googlemap(sel_gdirs)
# you might need to install motionless if it is not yet in your environment
Using OGGM#
OGGM can plot the thickness of a group of glaciers on a map:
graphics.plot_distributed_thickness(sel_gdirs)
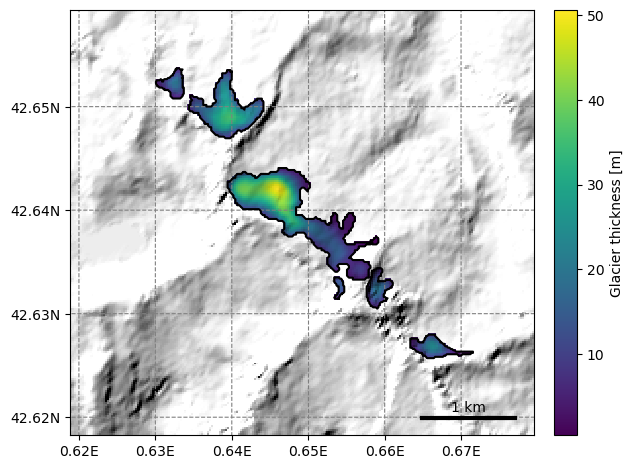
This is however not always very useful because OGGM can only plot on a map as large as the local glacier map of the first glacier in the list. See this issue for a discussion about why. In this case, we had a large enough border, and like that all neighboring glacers are visible.
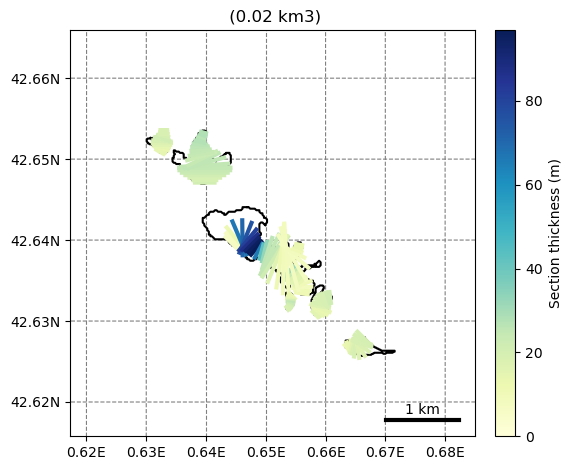
Since this issue, several glaciers can be plotted at once by the kwarg extend_plot_limit=True:
graphics.plot_inversion(sel_gdirs, extend_plot_limit=True)
Using salem#
Under the hood, OGGM uses salem to make the plots. Let’s do that for our case: it requires some manual tweaking, but it should be possible to automatize this better in the future.
Note: this also requires a version of salem after 21.05.2020
import salem
# Make a grid covering the desired map extent
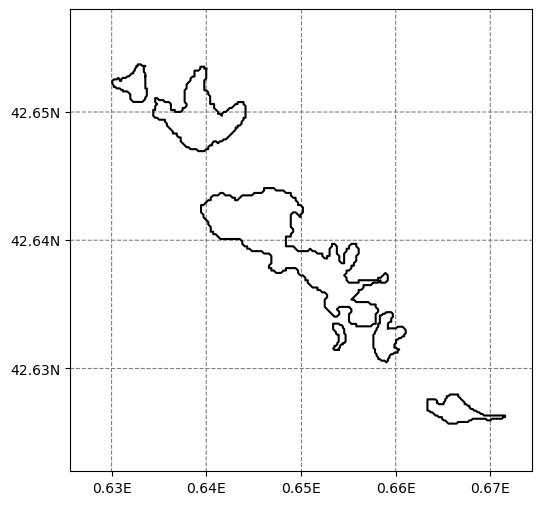
g = salem.mercator_grid(center_ll=(0.65, 42.64), extent=(4000, 4000))
# Create a map out of it
smap = salem.Map(g, countries=False)
# Add the glaciers outlines
for gdir in sel_gdirs:
crs = gdir.grid.center_grid
geom = gdir.read_pickle('geometries')
poly_pix = geom['polygon_pix']
smap.set_geometry(poly_pix, crs=crs, fc='none', zorder=2, linewidth=.2)
for l in poly_pix.interiors:
smap.set_geometry(l, crs=crs, color='black', linewidth=0.5)
f, ax = plt.subplots(figsize=(6, 6))
smap.visualize();
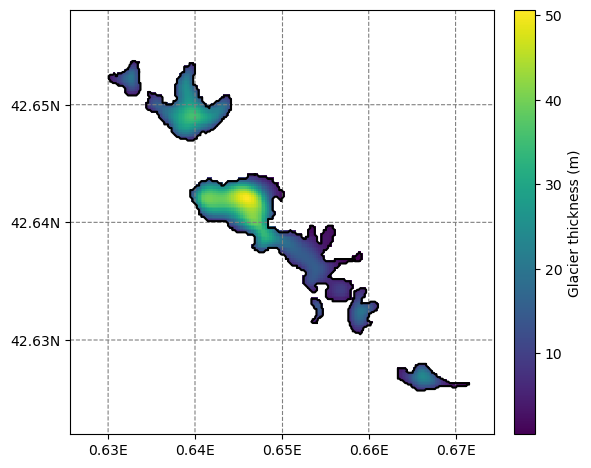
# Now add the thickness data
for gdir in sel_gdirs:
grids_file = gdir.get_filepath('gridded_data')
with utils.ncDataset(grids_file) as nc:
vn = 'distributed_thickness'
thick = nc.variables[vn][:]
mask = nc.variables['glacier_mask'][:]
thick = np.where(mask, thick, np.NaN)
# The "overplot=True" is key here
# this needs a recent version of salem to run properly
smap.set_data(thick, crs=gdir.grid.center_grid, overplot=True)
# Set colorscale and other things
smap.set_cmap(graphics.OGGM_CMAPS['glacier_thickness'])
smap.set_plot_params(nlevels=256)
# Plot
f, ax = plt.subplots(figsize=(6, 6))
smap.visualize(ax=ax, cbar_title='Glacier thickness (m)');
What’s next?#
return to the OGGM documentation
back to the table of contents


